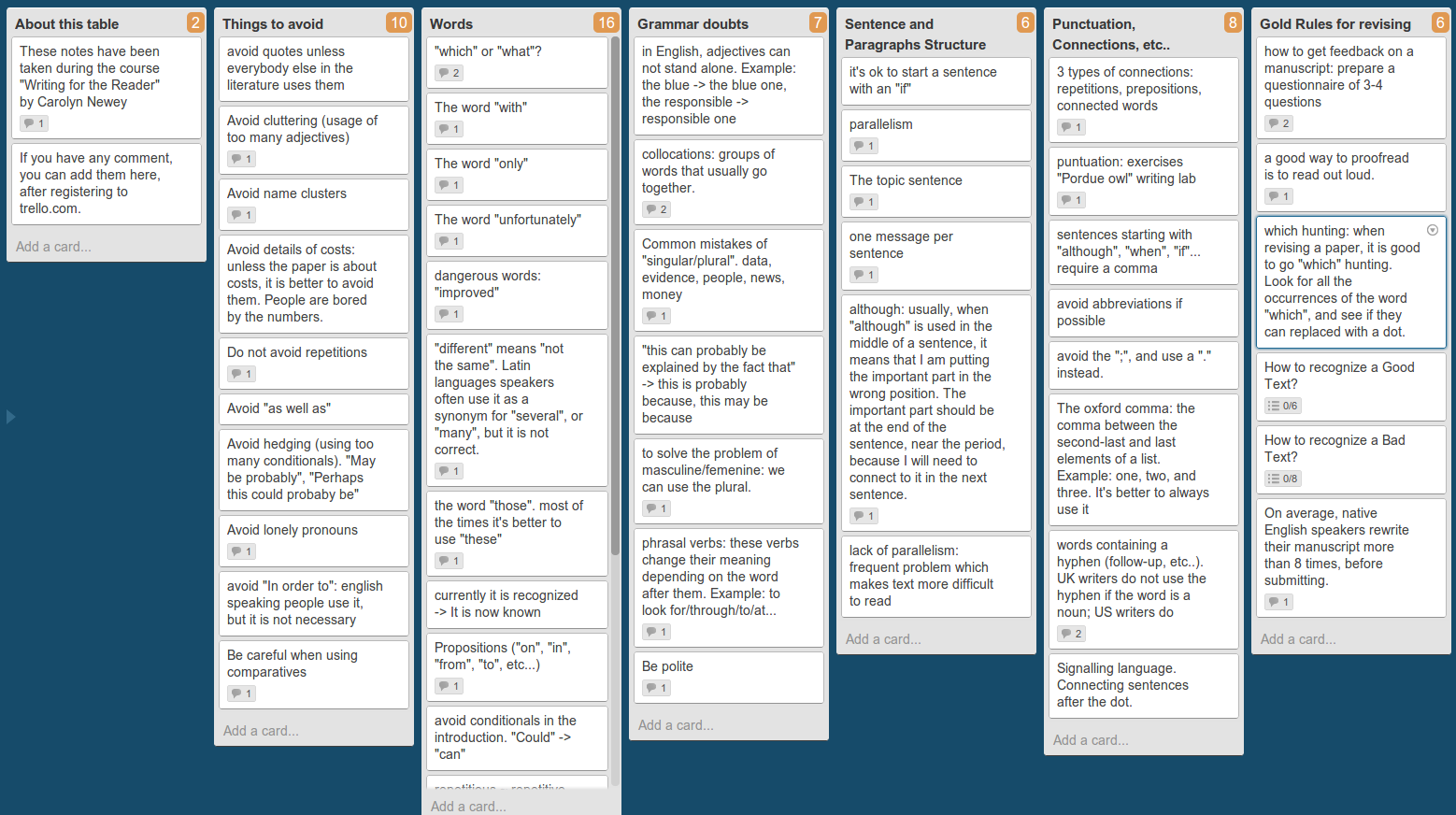
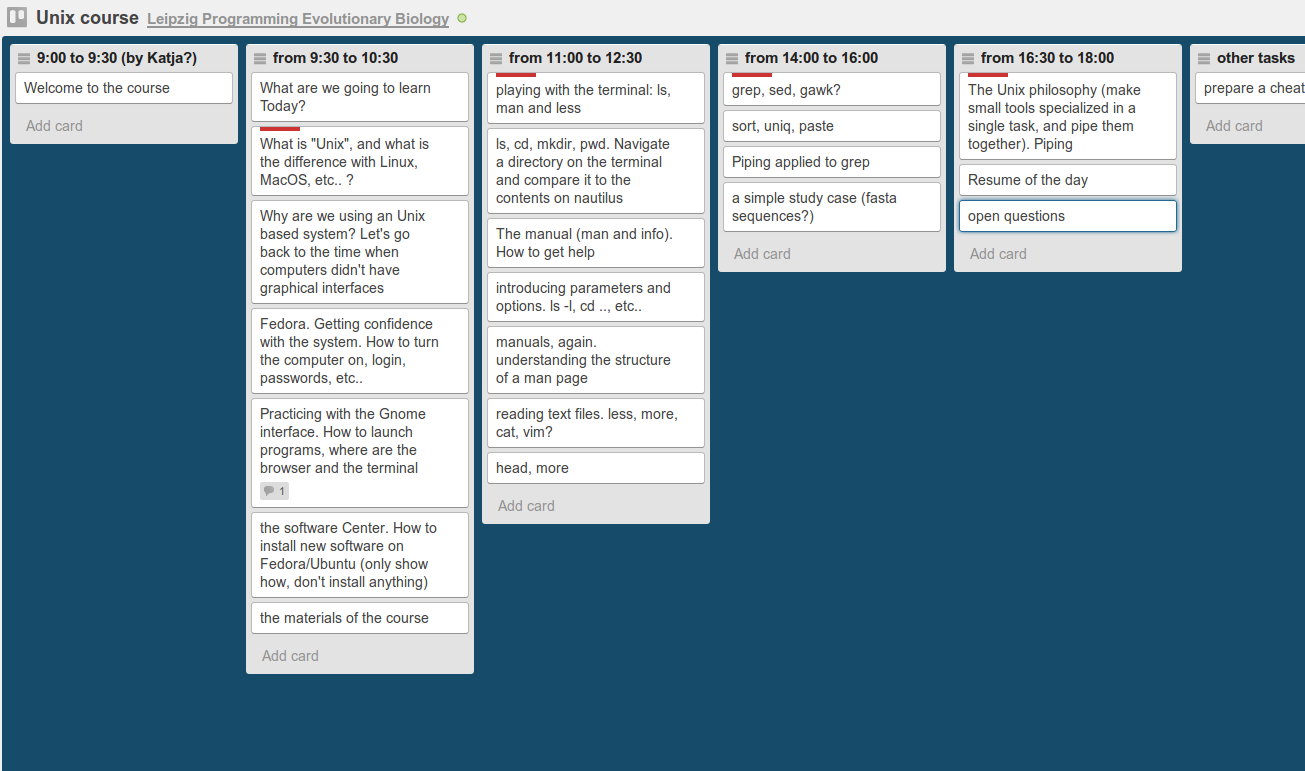
I have just come back from the Programming for Evolutionary Biology course in Leipzig, version 2013!! The course is still going on, but unfortunately this year I could not stay the whole duration three weeks, as I have stuff to do here in Barcelona.
This year, apart from the “Introduction to Linux” module, I also taught a short module on “Best Practices for programming in bioinformatics”. It was pure fun, I think I never enjoyed so much giving a talk. I explained a part about Version Control, and another about Scrum, and people were really excited about it. To make you understand how much people liked this talk, consider that three persons invited me a beer after that, which for me constitutes the maximum compliment for a talk.
I have uploaded the two slideshow on slideshare. Unfortunately, the best part of the talk was a live demonstration on how I use these practices during my daily work, but at the moment I can not make these example publicly available. However, you should be able to follow the slideshows anyway.